Alternative splicing induced by bacterial pore-forming toxins sharpens CIRBP-mediated cell response to Listeria infection
M. Corre, V. Böhm, V. Besic, A. Kurowska, A. Viry, A. Mohammad, C. Sénamaud-Beaufort, M. Thomas-Chollier, A. Lebreton
Authors :
Corre Morgane, Böhm Volker, Besic Vinko, Kurowska Anna, Viry Anouk, Mohammad Ammara, Sénamaud-Beaufort Catherine, Thomas-Chollier Morgane, Lebreton Alice
Abstract
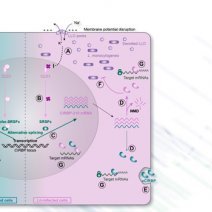
Cell autonomous responses to intracellular bacteria largely depend on reorganization of gene expression. To gain isoform-level resolution of these modes of regulation, we combined long- and short-read transcriptomic analyses of the response of intestinal epithelial cells to infection by the foodborne pathogen Listeria monocytogenes. Among the most striking isoform-based types of regulation, expression of the cellular stress response regulator CIRBP (cold-inducible RNA-binding protein) and of several SRSFs (serine/arginine-rich splicing factors) switched from canonical transcripts to nonsense-mediated decay-sensitive isoforms by inclusion of ‘poison exons’. We showed that damage to host cell membranes caused by bacterial pore-forming toxins (listeriolysin O, perfringolysin, streptolysin or aerolysin) led to the dephosphorylation of SRSFs via the inhibition of the kinase activity of CLK1, thereby driving CIRBP alternative splicing. CIRBP isoform usage was found to have consequences on infection, since selective repression of canonical CIRBP reduced intracellular bacterial load while that of the poison exon-containing isoform exacerbated it. Consistently, CIRBP-bound mRNAs were shifted towards stress-relevant transcripts in infected cells, with increased mRNA levels or reduced translation efficiency for some targets. Our results thus generalize the alternative splicing of CIRBP and SRSFs as a common response to biotic or abiotic stresses by extending its relevance to the context of bacterial infection.
Nucleic Acids Research. 2023 Nov 6 ; gkad1033 ; doi:10.1093/nar/gkad1033