![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | SRR576933.fastq |
| File type | Conventional base calls |
| Encoding | Illumina 1.5 |
| Total Sequences | 3603544 |
| Filtered Sequences | 0 |
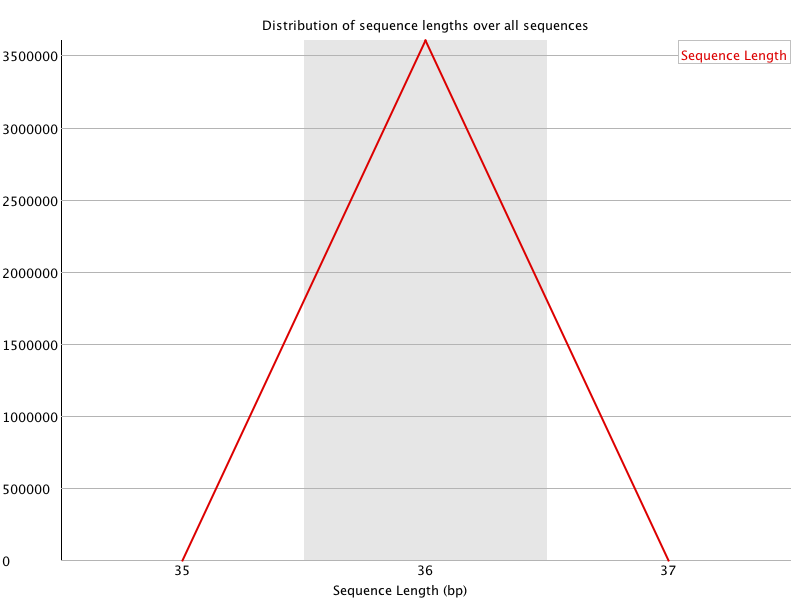
| Sequence length | 36 |
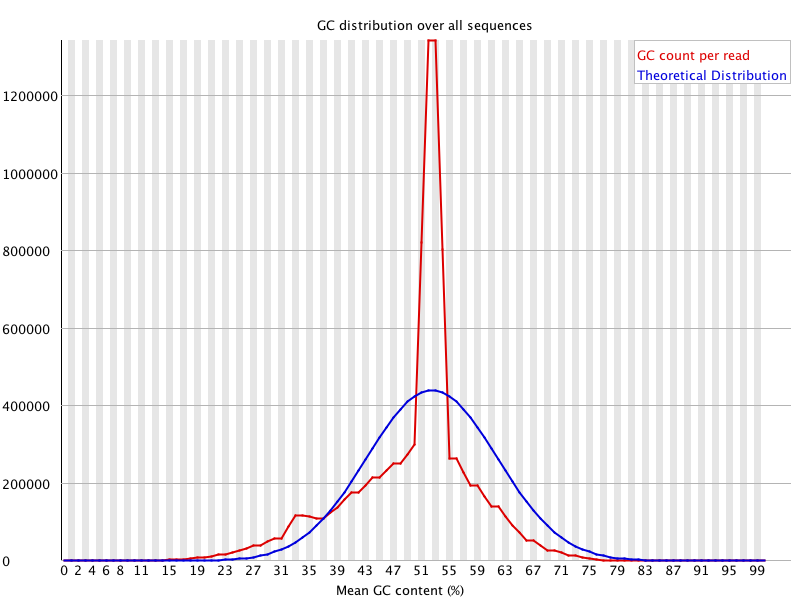
| %GC | 49 |
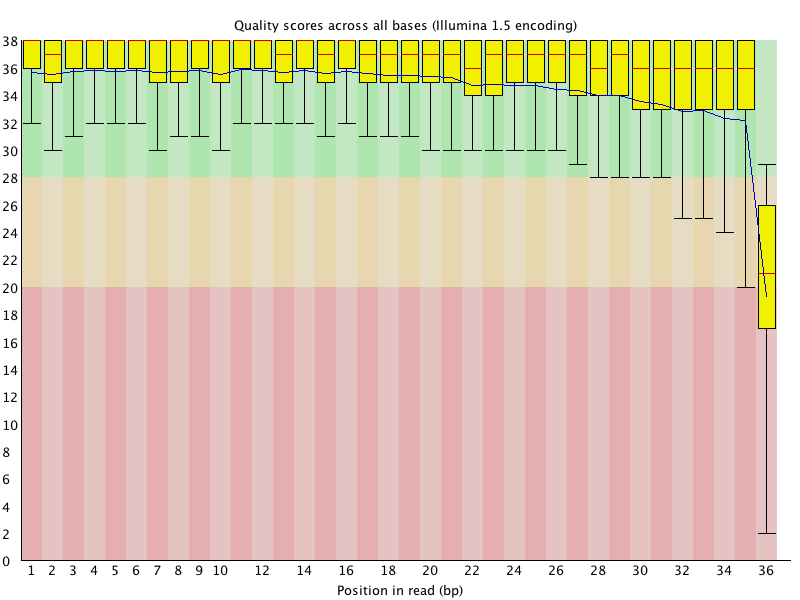
![[WARN]](Icons/warning.png) Per base sequence quality
Per base sequence quality
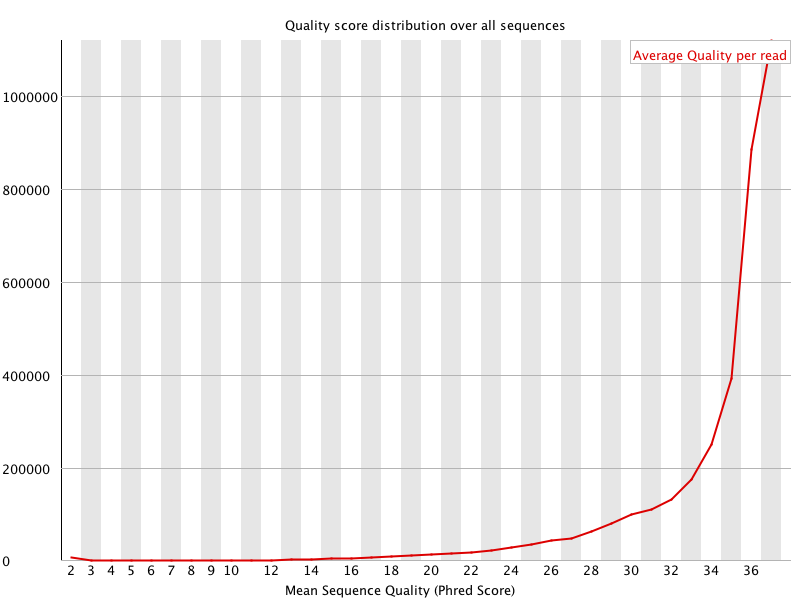
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
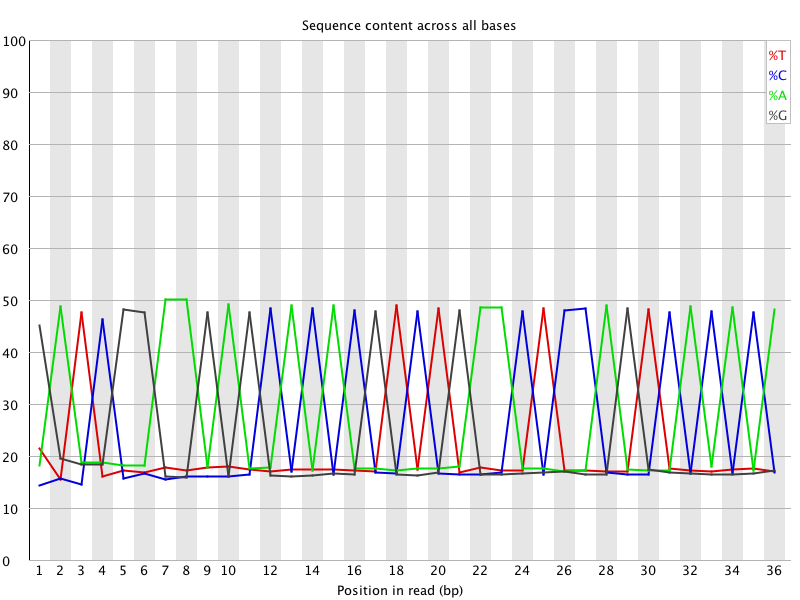
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
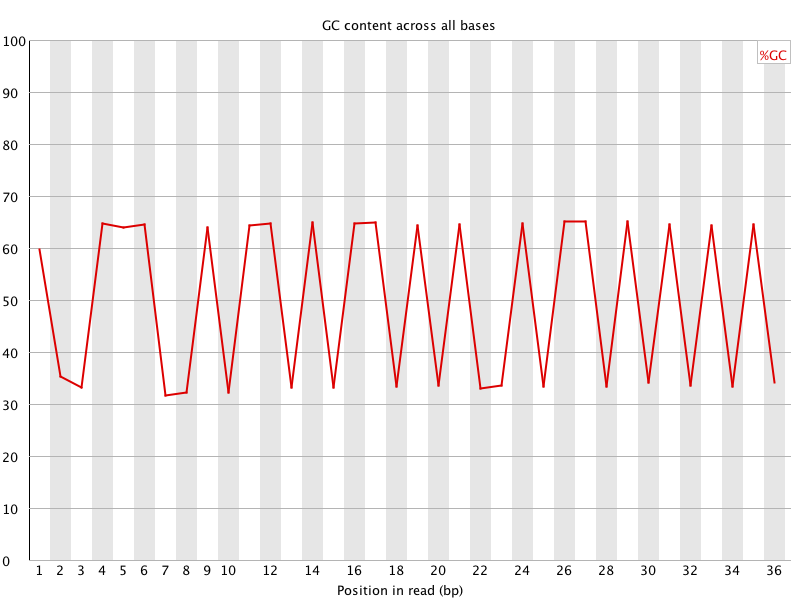
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content

![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
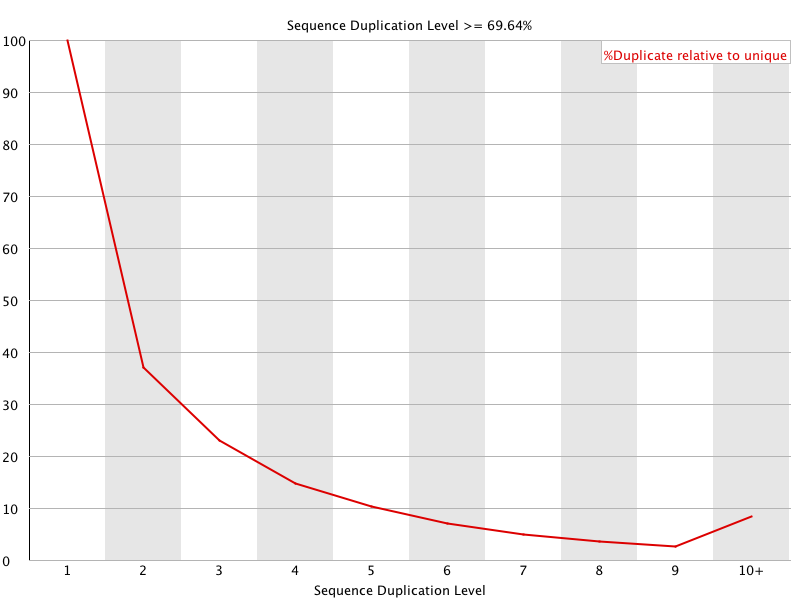
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[FAIL]](Icons/error.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACACA | 1060621 | 29.432719567181643 | TruSeq Adapter, Index 5 (100% over 36bp) |
| GCTAACAAATACCCGACTAAATCAGTCAAGTAAATA | 13630 | 0.37823875606902535 | No Hit |
| NATCGGAAGAGCACACGTCTGAACTCCAGTCACACA | 11728 | 0.3254573830651159 | TruSeq Adapter, Index 5 (97% over 36bp) |
| GTTAGCTATTTACTTGACTGATTTAGTCGGGTATTT | 10983 | 0.304783291115635 | No Hit |
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACACC | 3658 | 0.10151117899490057 | TruSeq Adapter, Index 1 (97% over 36bp) |
![[FAIL]](Icons/error.png) Kmer Content
Kmer Content
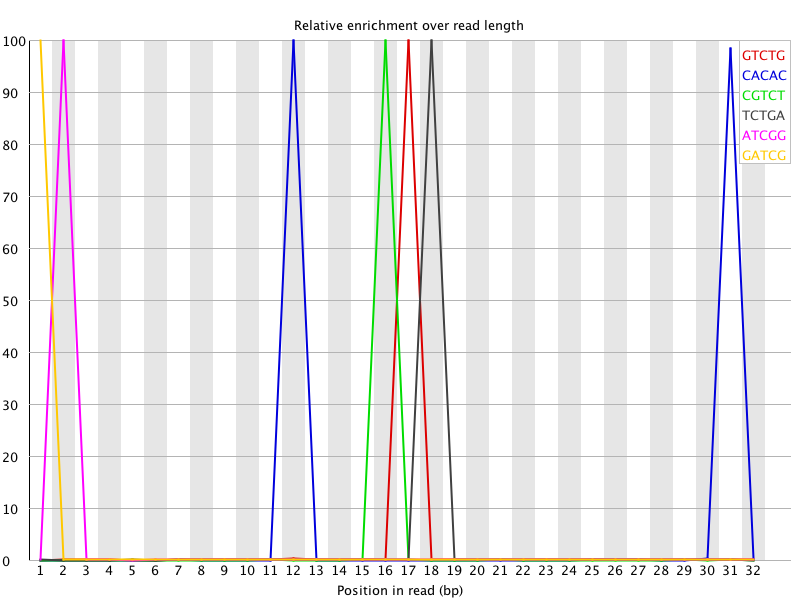
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GTCTG | 1221940 | 15.112505 | 449.36426 | 17 |
| CACAC | 2314305 | 14.062944 | 220.92882 | 12 |
| CGTCT | 1196835 | 13.471048 | 409.01773 | 16 |
| TCTGA | 1211805 | 12.584683 | 376.985 | 18 |
| ATCGG | 1218190 | 11.603947 | 346.49387 | 2 |
| GATCG | 1211235 | 11.537695 | 347.40747 | 1 |
| TCGGA | 1176255 | 11.204492 | 346.44058 | 3 |
| TCCAG | 1248195 | 10.820656 | 313.0933 | 25 |
| CCAGT | 1237425 | 10.72729 | 312.60504 | 26 |
| CAGTC | 1204935 | 10.445633 | 312.02316 | 27 |
| ACGTC | 1198605 | 10.3907585 | 315.23105 | 15 |
| GTCAC | 1190165 | 10.317591 | 312.2194 | 29 |
| CACGT | 1186765 | 10.288116 | 315.0924 | 14 |
| CTGAA | 1268955 | 10.149855 | 290.48465 | 19 |
| GGAAG | 1237160 | 9.973329 | 294.04337 | 5 |
| TGAAC | 1225130 | 9.799316 | 289.98593 | 20 |
| AGTCA | 1204960 | 9.637985 | 288.0274 | 28 |
| GAACT | 1201780 | 9.61255 | 289.62643 | 21 |
| CTCCA | 1177075 | 9.286579 | 284.30338 | 24 |
| ACTCC | 1171410 | 9.241885 | 284.8321 | 23 |
| CGGAA | 1236915 | 9.074748 | 267.3231 | 4 |
| GAGCA | 1219600 | 8.947715 | 267.08813 | 9 |
| AGAGC | 1197790 | 8.7877035 | 266.93942 | 8 |
| AACTC | 1190695 | 8.667517 | 263.18796 | 22 |
| TCACA | 1184355 | 8.621364 | 260.89795 | 30 |
| GAAGA | 1259060 | 8.522827 | 247.18573 | 6 |
| AAGAG | 1215850 | 8.2303295 | 246.49074 | 7 |
| AGCAC | 1215760 | 8.117514 | 242.63748 | 10 |
| GCACA | 1209255 | 8.074081 | 242.5414 | 11 |
| ACACG | 1186520 | 7.9222813 | 242.69942 | 13 |
| ACACA | 1158640 | 6.49601 | 199.66386 | 32 |
| TTTTT | 234880 | 4.13627 | 4.749043 | 31 |
| TATTT | 158625 | 2.151486 | 7.856395 | 7 |
| TGATT | 150440 | 1.8715926 | 6.969653 | 19 |
| GATTT | 150010 | 1.8662431 | 6.9205275 | 20 |
| CTGAT | 151850 | 1.5769733 | 5.816326 | 18 |
| ATTTA | 146225 | 1.52754 | 5.7619843 | 8 |
| TTTAC | 125675 | 1.4229099 | 6.2630315 | 9 |
| GTATT | 103770 | 1.2909809 | 5.408579 | 31 |
| TCAGT | 100520 | 1.0439075 | 5.499591 | 22 |
| TTACT | 88195 | 0.99855596 | 5.6727595 | 10 |
| GTCGG | 79730 | 0.90446025 | 5.540607 | 26 |
| TTTAG | 69825 | 0.86867833 | 5.935576 | 22 |
| GCTAT | 82965 | 0.8615976 | 5.2448087 | 5 |
| TTGAC | 80810 | 0.83921784 | 5.1202207 | 14 |
| TTAGC | 78895 | 0.81933033 | 5.1235423 | 2 |
| TACTT | 69795 | 0.79022866 | 5.5242867 | 11 |
| CTATT | 69415 | 0.7859262 | 5.575016 | 6 |
| TGACT | 75445 | 0.78350186 | 5.0687356 | 15 |
| TCGGG | 67285 | 0.7632838 | 5.3157706 | 27 |
| TTAGT | 57975 | 0.7212549 | 5.807995 | 23 |
| GTTAG | 59300 | 0.6766807 | 5.3769717 | 1 |